Sulfur (S) is one of the essential elements for plants.
Plants produce cysteine and methionine, an essential amino acid for humans, from sulfate in the environment. Plants also play a significant role in the S cycle in nature by transforming inorganic S into organic compounds. S-containing compounds play pivotal roles in the metabolic process by working as redox substances, coenzymes, and signaling molecules. Glucosinolates in cruciferous plants have anti-carcinogenic activity for humans.
We are trying to understand how plants regulate S assimilation and metabolism in response to various environmental factors to provide clues for improving crop productivity, promoting human health, and solving environmental problems.
Sulfur Assimilation and Metabolism
Sulfur (S) assimilation starts from sulfate uptake, facilitated by sulfate transporters (SULTR). Then sulfate is activated and reduced by several enzymes and assimilated into cysteine (Review 11). S-containing metabolites, including amino acids methionine, major reductant glutathione, and specific metabolites, are synthesized from cysteine.
When the environmental S is limited, expression of S assimilatory genes increases, and glucosinolates' biosynthesis is repressed (Article 11, Review 9). These responses to S limitation support plant life in the environment.
We are trying to elucidate the mechanisms of S assimilation and metabolism and their regulatory mechanisms by analyzing the plant responses to S limitations.

Mechanism of Sulfur Deficiency Responses
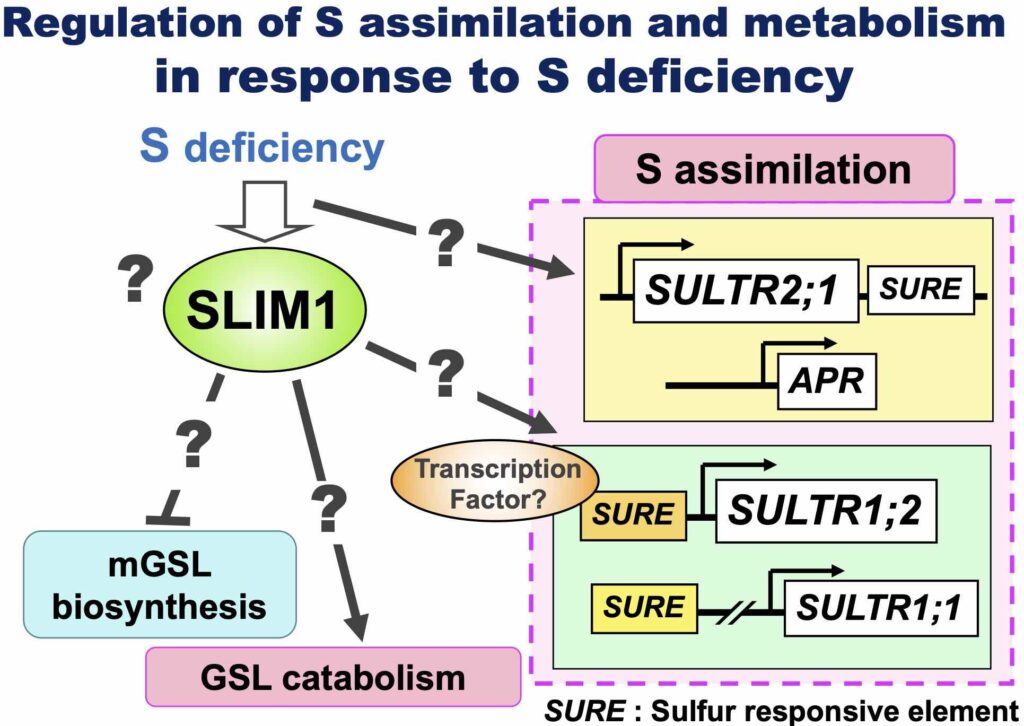
How do plants sense S deficiency and regulate their metabolism?
We identified SLIM1, a central transcriptional regulator responsible for increased S assimilation and repression of glucosinolates biosynthesis under S deficiency (Article 16).
Still, there are many questions remain, such as "How do plants sense S deficiency?", "How does SLIM1 work?" etc. We will find a way to activate S assimilation by solving these questions.
Regulation of Sulfate Uptake
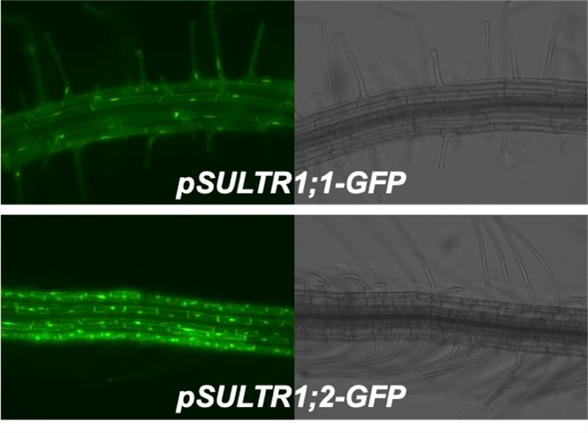
In Arabidopsis, the sulfate transporters SULTR1;1 and SULTR1;2 are responsible for sulfate uptake.
S deficiency stimulates their expression and increases sulfate uptake activity.
We have determined the regulatory sequences responsible for the upregulation of SULTR1;1 (Article 15). We also found that phosphorylation and dephosphorylation of proteins are involved in this response (Article 12).
SULTR1;1 and SULTR1;2 work in the root surface cell layers, such as the epidermis and cortex. The epidermis consists of root hair cells and non-hair cells where SULTR1;1 and SULTR1;2 are separately expressed (Article 32). We are interested in the mechanism and physiological meaning of this beautiful separation.
Regulatory Mechanisms of Biosynthesis and Metabolism of Aliphatic Glucosinolates
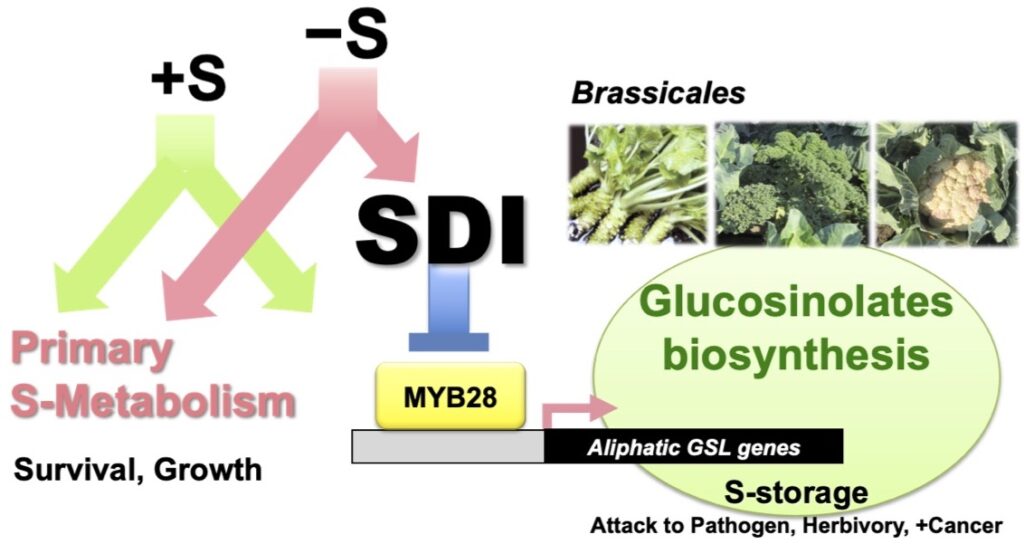
Glucosinolates (GSL), unique S-containing metabolites in cruciferous plants, act as repellents to pesticides and insects and attract certain insects. It also functions in S storage. Some methionine-derived aliphatic GSL (mGSLs) show anti-carcinogenic activity.
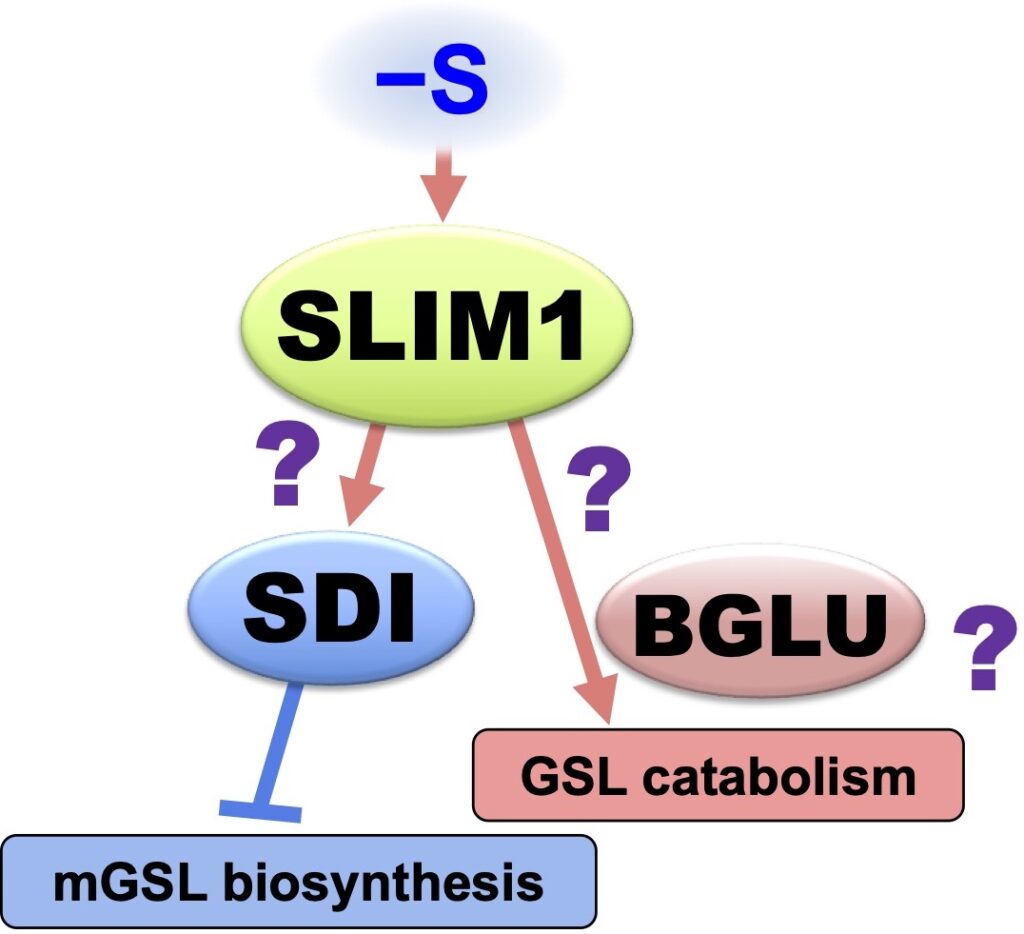
Sulfur Deficiency Induced (SDI) proteins suppress mGSL bio-synthesis. S deficiency activates SDI1 expression, and SDI1 protein interacts with MYB28, a transcription factor that promotes mGSL biosynthesis, thereby suppressing its function and mGSL biosynthesis gene expression. This process eventually suppresses mGSL bio-synthesis under S deficiency (Article 26).
ß-Glucosidase (BGLU), called myrosinase, is responsible for GSL catabolism. S deficiency activates gene expression of BGLU28 and BGLU30 (Article 11, 15, 16). We found that they are responsible for GSL catabolism under S deficiency, and this process sustains plant life by providing sulfate to S assimilation under S limitation (Article 34).
Novel Mechanisms Controlling S Assimilation and Metabolism
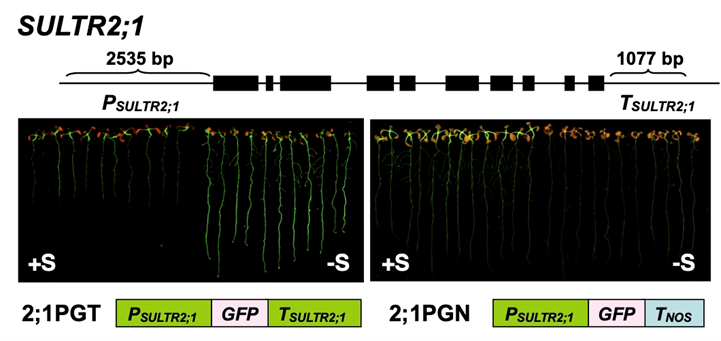
SULTR2;1 is a representative gene of which S deficiency response is independent of SLIM1. We identified the S deficiency responsive elements in the 3' region of SULTR2;1 (Article 23). Further clarification of SULTR2;1 gene expression would provide a novel regulatory mechanism of S assimilation.
Influences of S on other elements' distribution and metabolism
S distribution and metabolism are mutually regulated with those of other nutrients. The nitrogen and S ratio in organisms is maintained, and carbon and nitrogen levels influence the S deficiency response (Article 14).
S deficiency stimulates phosphate uptake and distribution to shoots (Article 38). Vice versa, phosphorus deficiency alters sulfate distribution. A similar relationship exists between S and iron. We are interested in the mechanisms of such mutual regulation among nutrients and the physiological meaning in plants.
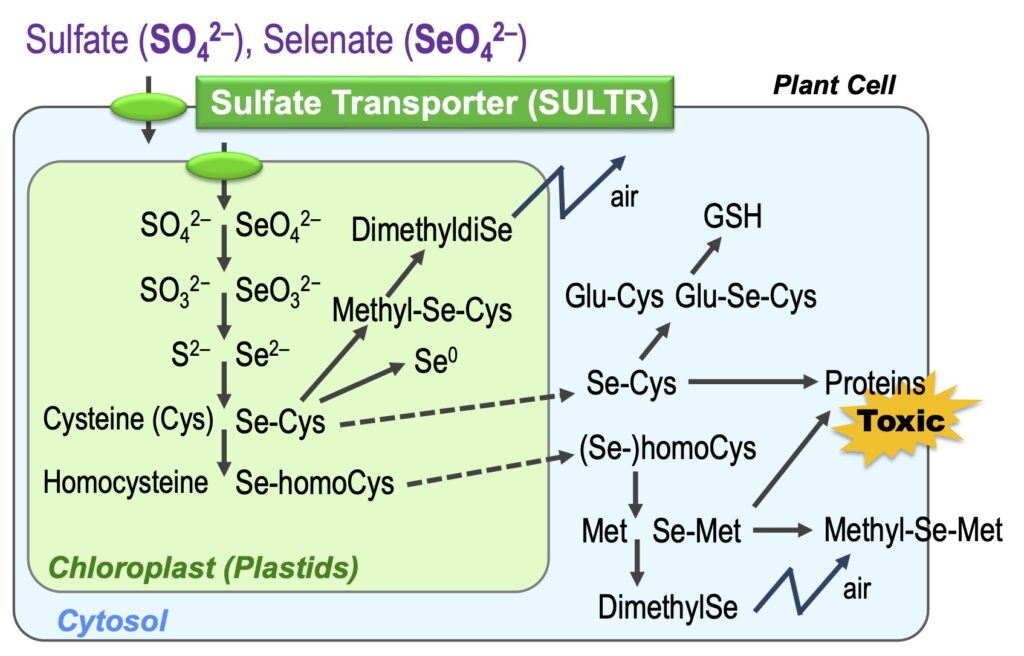
Selenium (Se) is a family element of S and is metabolized similarly in the plants. Se is an essential element for animals and micro-organisms and becomes toxic at higher doses. We are trying to clarify the role of Se in higher plants and the mechanisms of its toxicity.
We developed a monitoring system for selenate and chromate in the environment using SULTR promoter-GFP plants (Article 17,25).
S in Abiotic and Biotic Stress
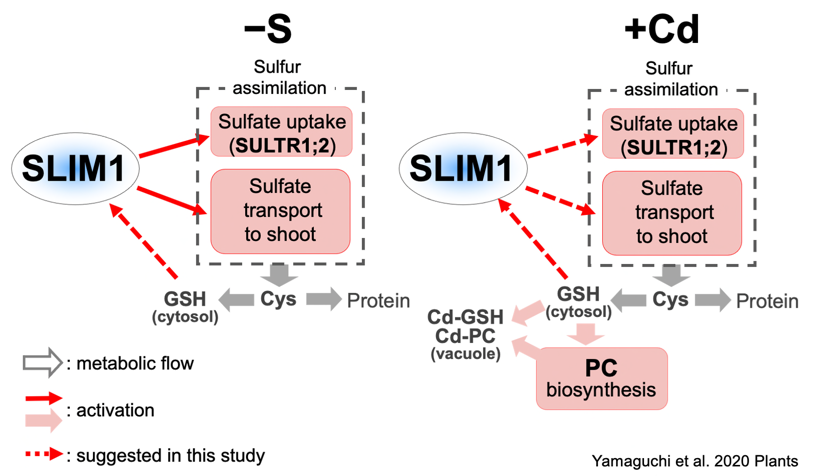
S is closely related to stress tolerance in plants. Glutathione and phytochelatins detoxify heavy metals through S moiety. S assimilation and phytochelatin synthesis are enhanced in plants exposed to heavy metals.
Sulfate transport to shoots is also stimulated by heavy metals (Article 27). Upon heavy metal stress, phytochelatin synthesis is prioritized even in the absence of S. SULTR1;2 and SLIM1 contribute to these responses (Article 27, 29, 35).
The contribution of S metabolism to biotic stress is also an interesting topic (Article 20, 37).
Breeding plants with higher S assimilation and beneficial S-containing compounds
Soybean is an important source of plant-derived protein but is limited to S-containing amino acids methionine. So, methionine is added to livestock feed. We are attempting to increase the amount of S in the edible parts of plants using our research outcomes.
Chemicals and environmental factors influence S assimilation and metabolism
The discovery of environmental factors and compounds that alter sulfate uptake and assimilation will help cultivate ways to maintain higher S assimilation efficiencies.
We found that plant hormone cytokinin inhibits sulfate uptake (Article 13), γ-aminolevulinic acid increases sulfate uptake and cysteine and glutathione levels, and low salt concentrations promote plant growth S accumulation (Article 41, 22).
Finding other compounds and environmental factors that improve S assimilation will be the clues to elucidate the regulatory system and provide helpful knowledge for agriculture.
